Asa’s R packages 📦
for bioinformatics and data analysis
Chen Peng
ZJU LSI Jianglab
2024-04-19
My R packges
My favorite programming language is R, I have developed some R packages:

pcutils 
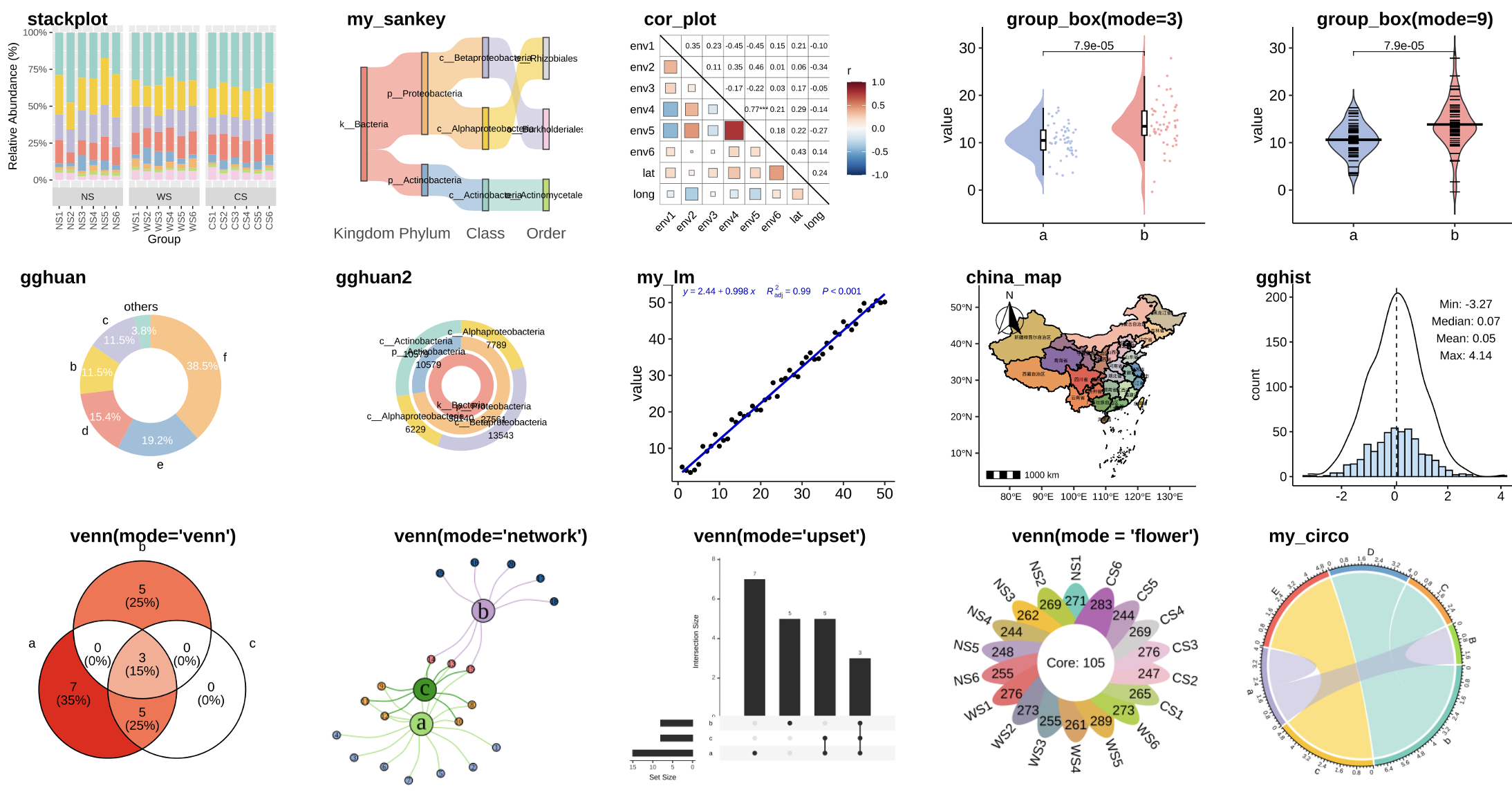
It contains many useful functions for statistics and visualization.
Go down 👇
pctax 
- It contains a basic pipeline for omics analysis.
- See the pctax Tutorial

pctax features
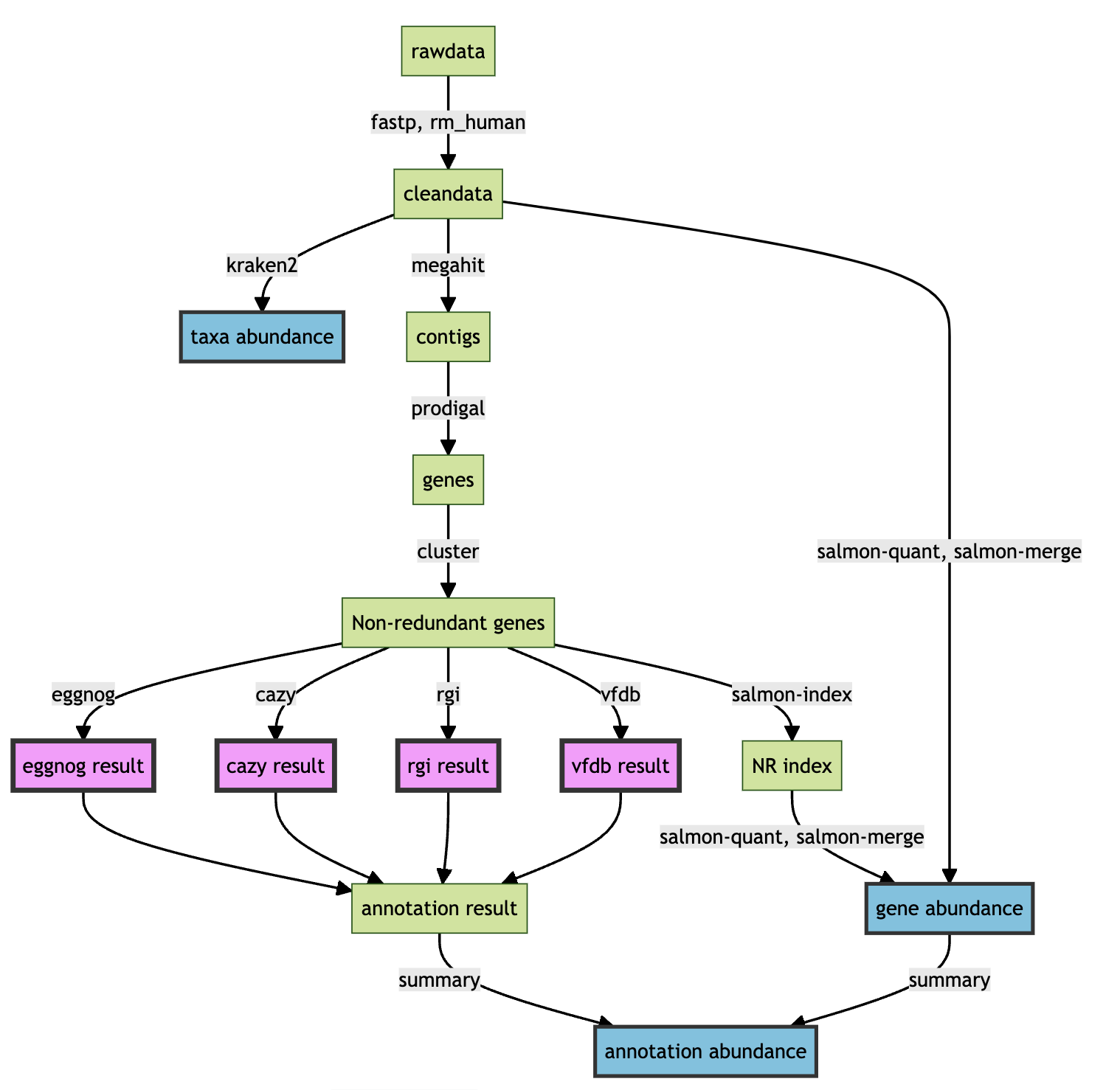
Built-in steps in the micro_sbatch():
- fastp
- rm_human
- kraken2
- megahit
- prodigal
- cluster
- salmon-quant
- eggnog
- cazy
- rgi
- vfdb
- …
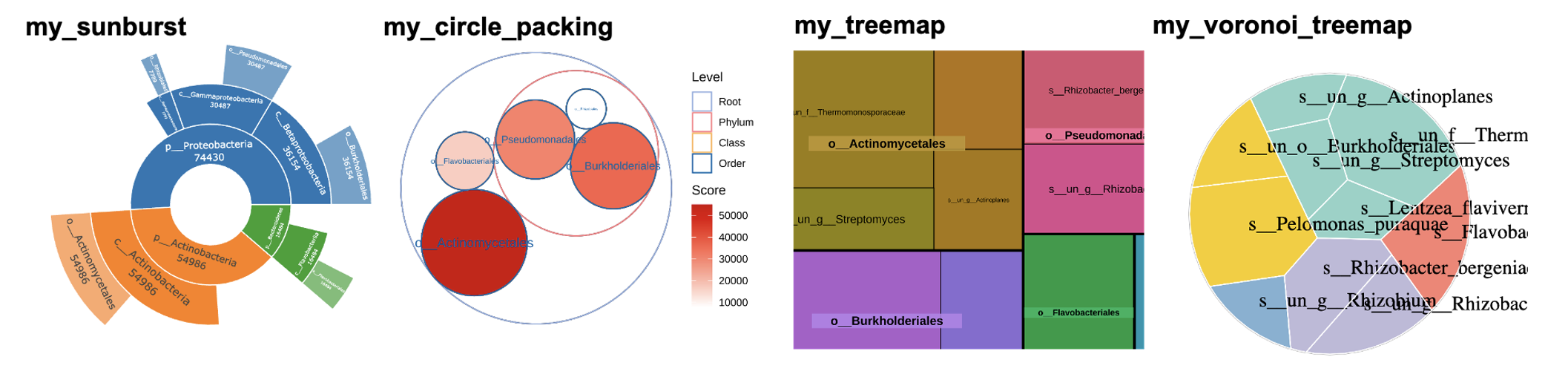
Taxonkit is a Practical and Efficient NCBI Taxonomy Toolkit.
You can use Taxonkit in R by pctax interface as followed:
taxid Kingdom Phylum Class
1 562 k__Bacteria p__Pseudomonadota c__Gammaproteobacteria
2 9606 k__Eukaryota p__Chordata c__Mammalia
Order Family Genus Species
1 o__Enterobacterales f__Enterobacteriaceae g__Escherichia s__Escherichia coli
2 o__Primates f__Hominidae g__Homo s__Homo sapiensUnified input and output format of one type of analysis for convenient downstream
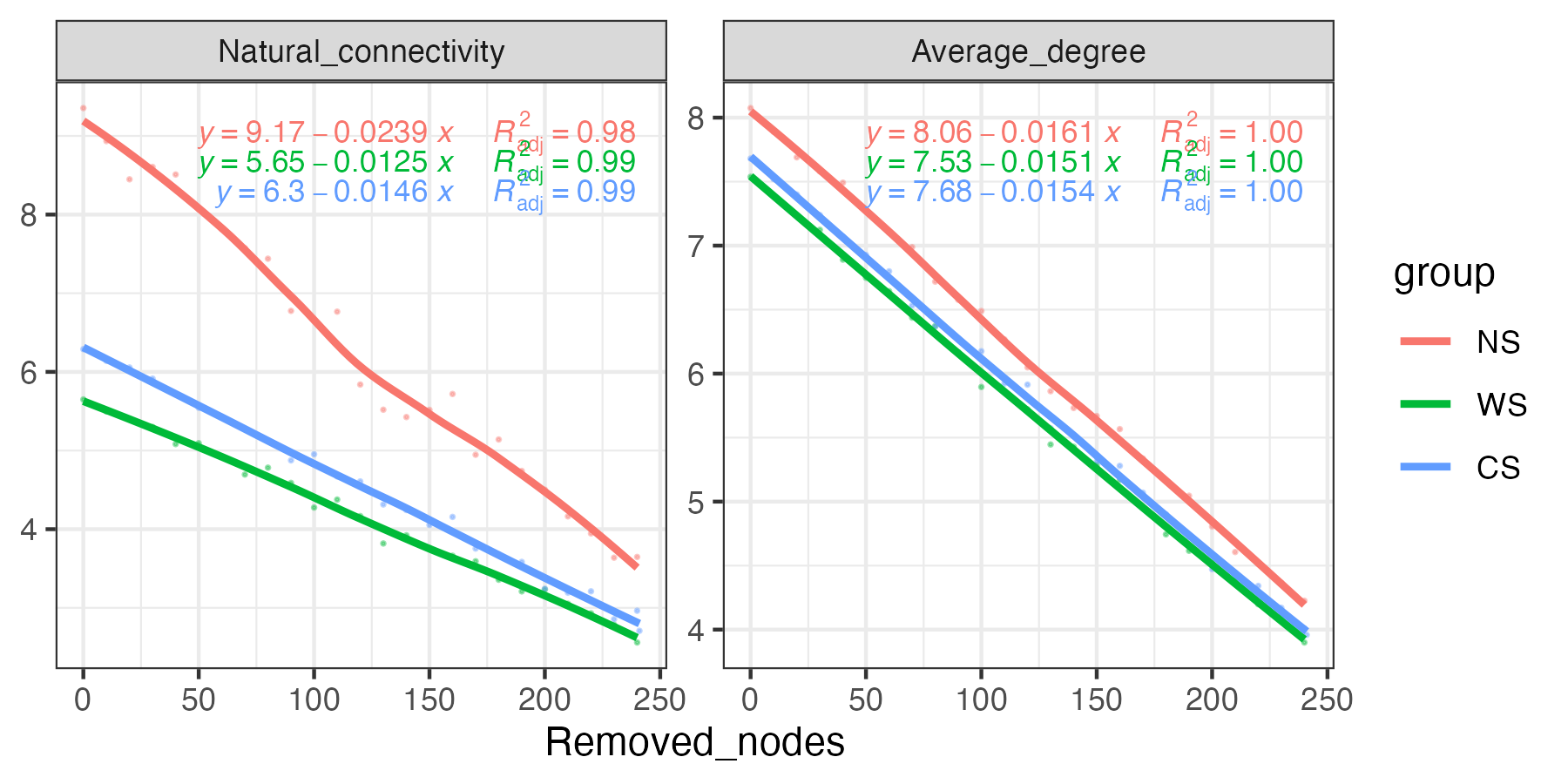
- α-diversity
- Richness, Shannon, Simpson, Chao1, ACE, PD, Pielou, …
- β-diversity
- Distance: Bray-Curtis, Jaccard, Unifrac, Beta MPD, Phylosor, …
- Ordination: PCA, PCoA, NMDS, PLS-DA, t-SNE, UMAP, LDA, RDA, CCA, db-RDA, …
- PERMANOVA: adonis, anosim, mrpp, mantel, envfit, …
- ζ-diversity
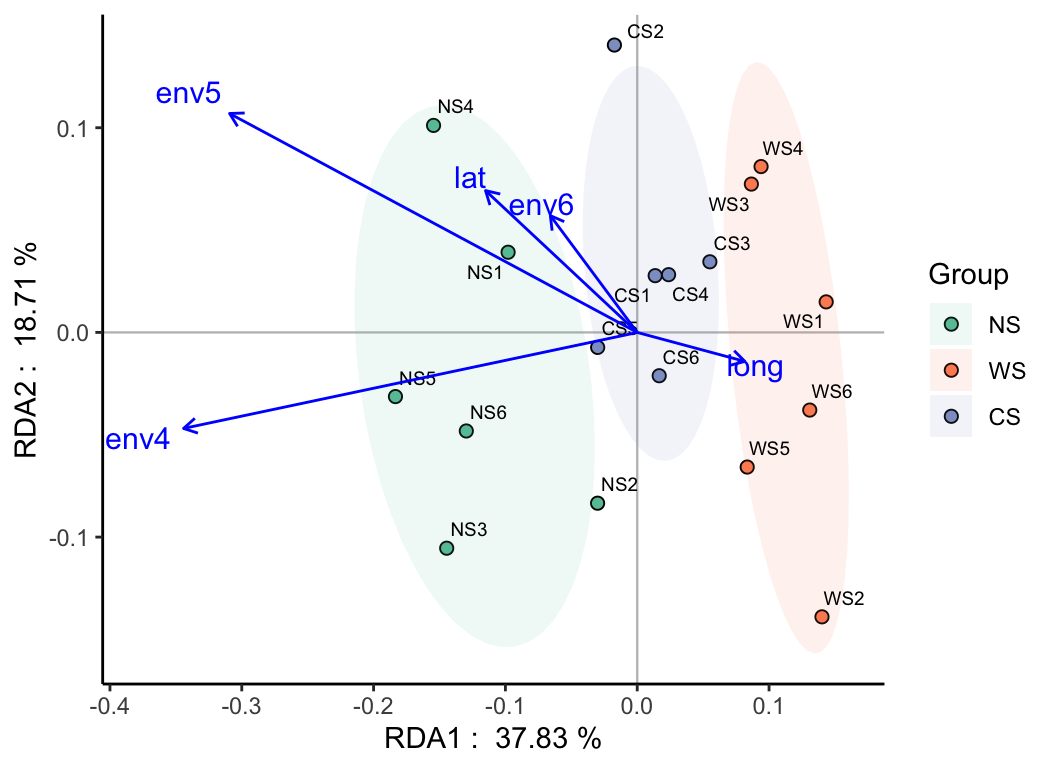
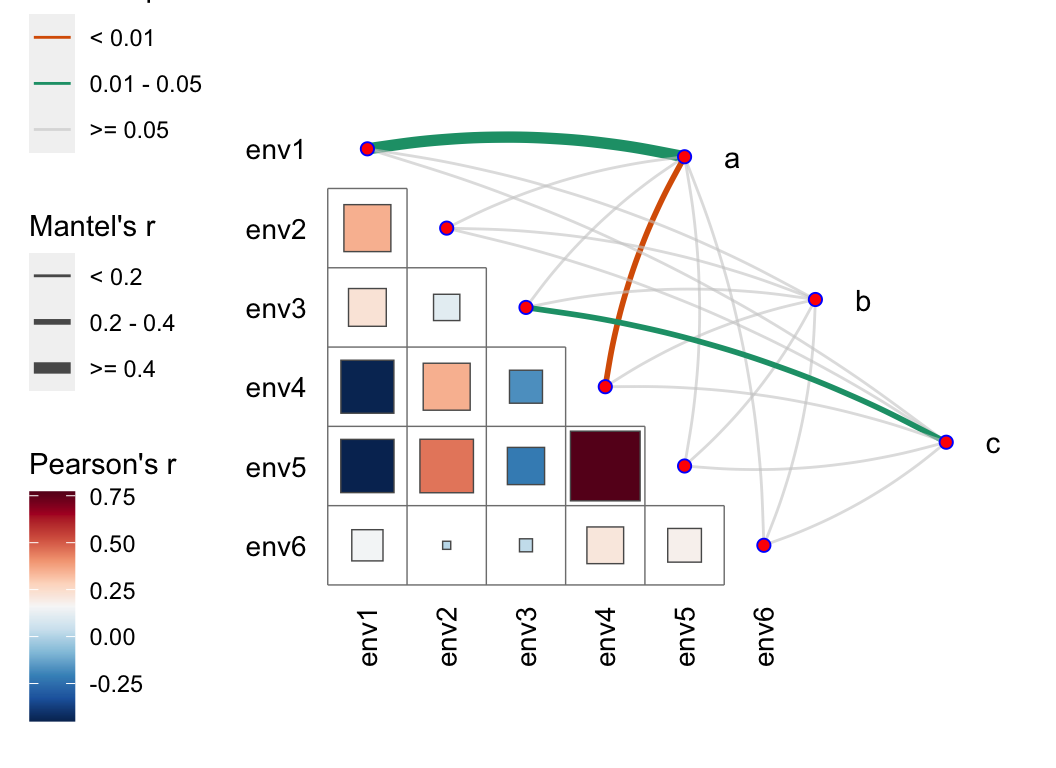
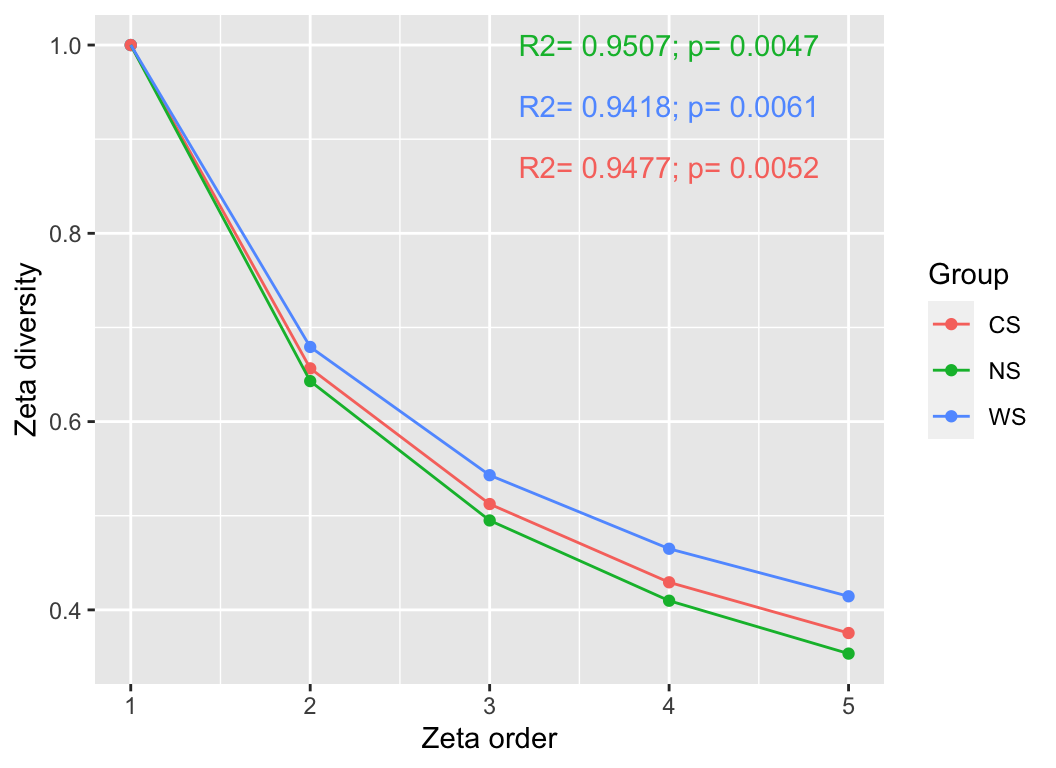
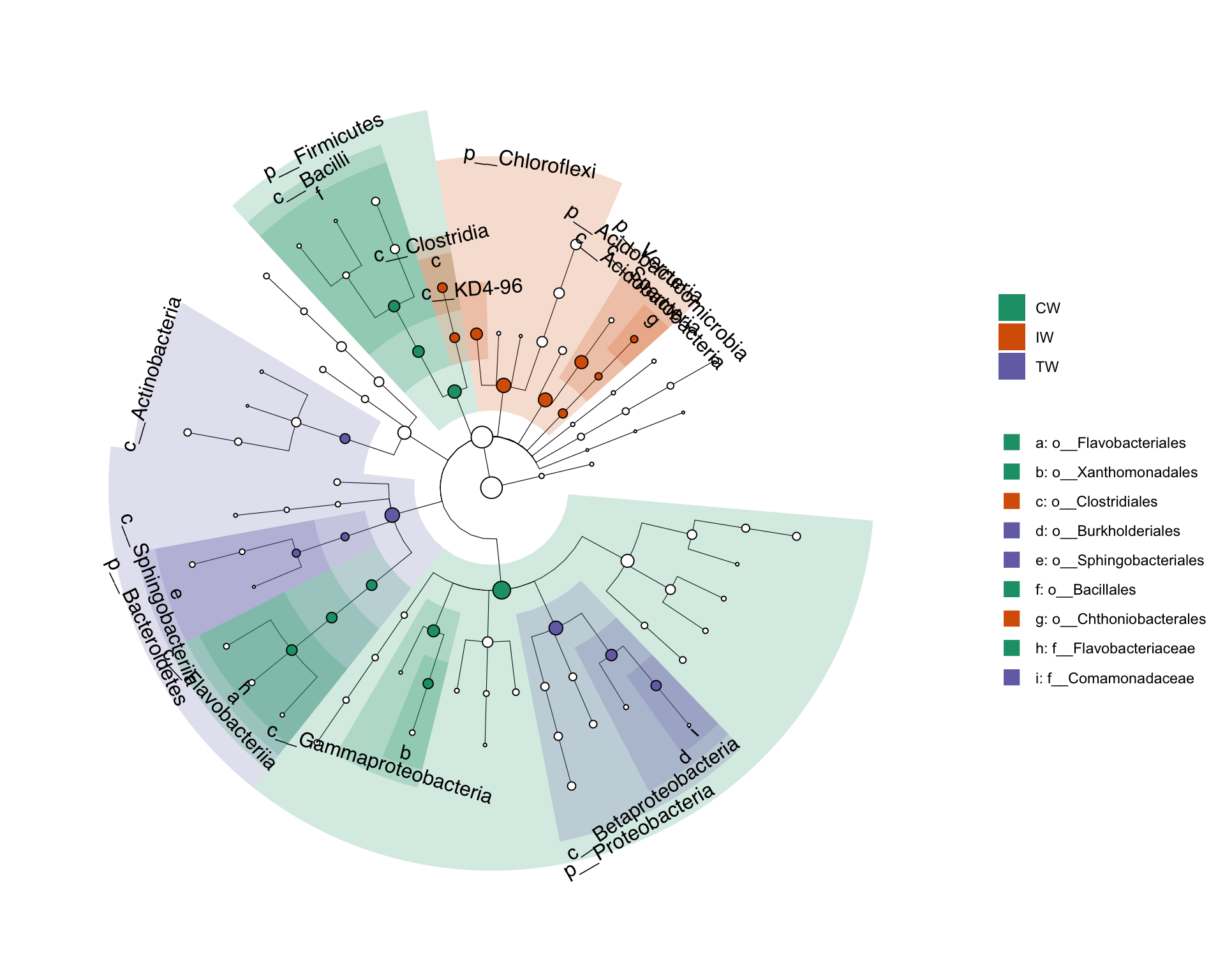
pctax features
- t.test
- wilcox.test
- DESeq2
- edgeR
- limma
- randomForest
- LEfSe
- …
- Solan NCM
- Stegen (βNTI & RCbray-based)
- ST, NST, MST (stochasticity ratio)
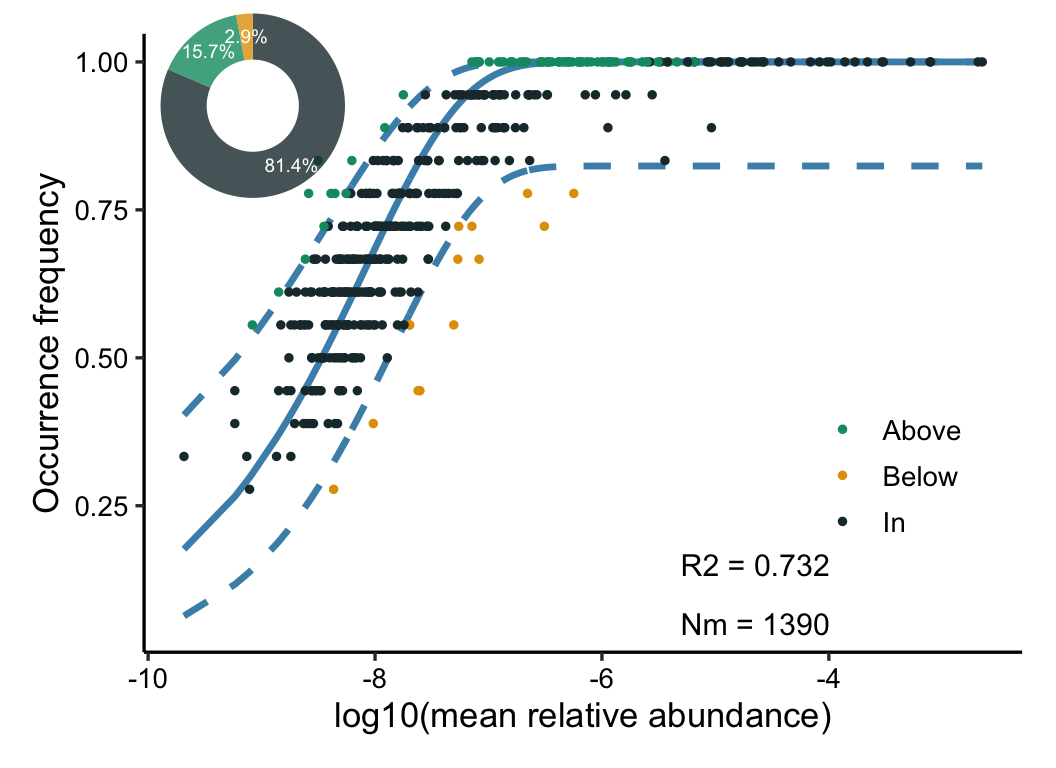
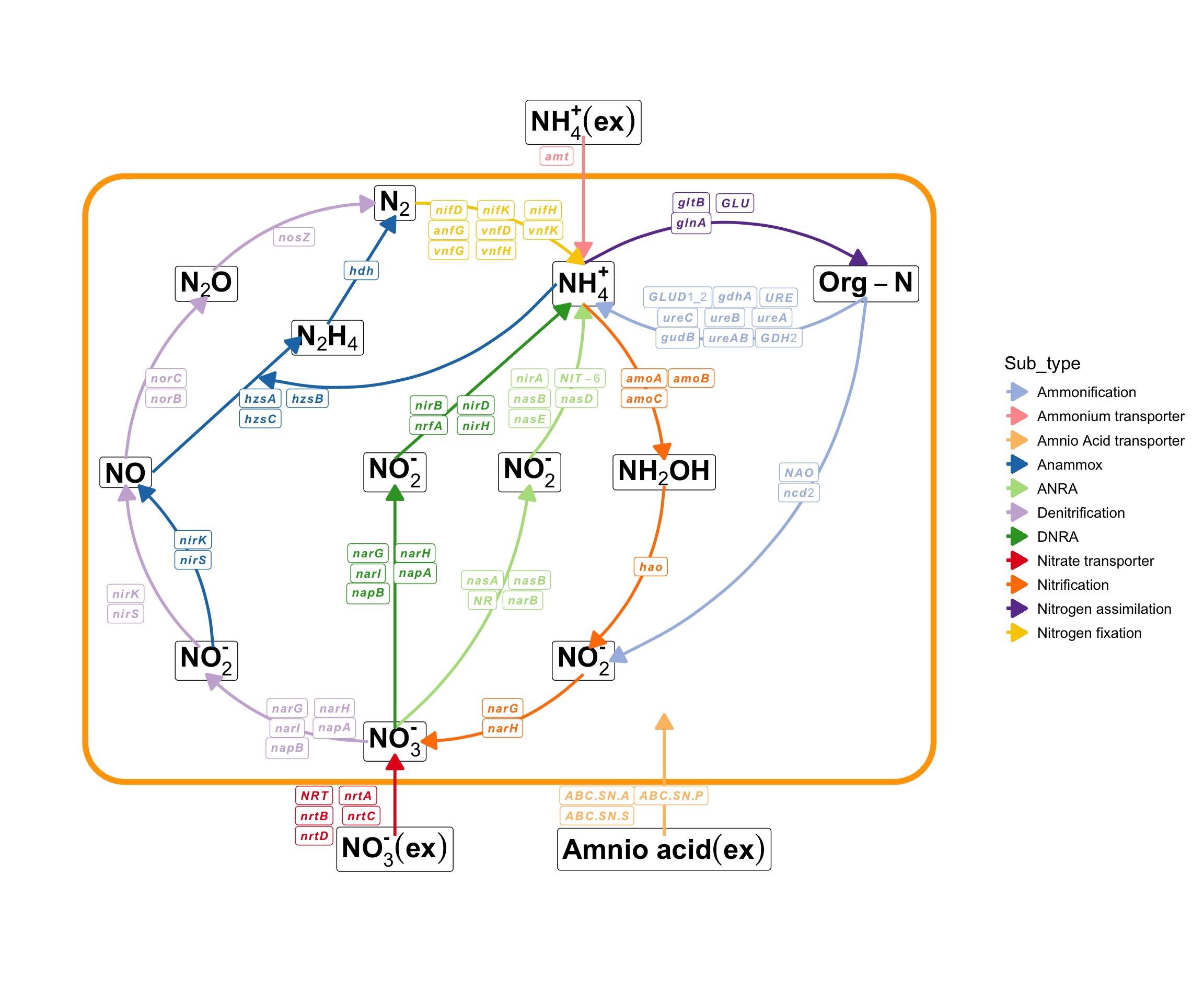
The pctax package includes some built-in cycling pathways, which enables users to conveniently visualize all genes or KO (KEGG Orthology) involved in these pathways.
MetaNet 
- It is a comprehensive network analysis package for omics data.
- See the MetaNet Tutorial

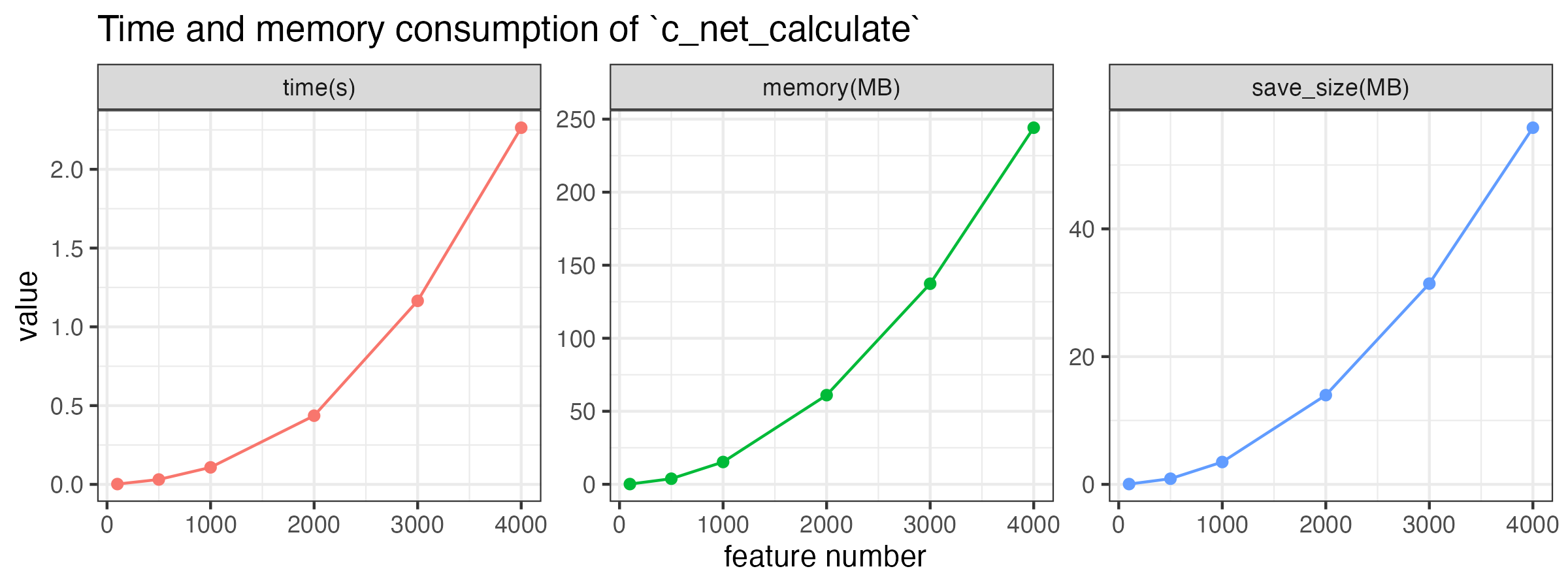
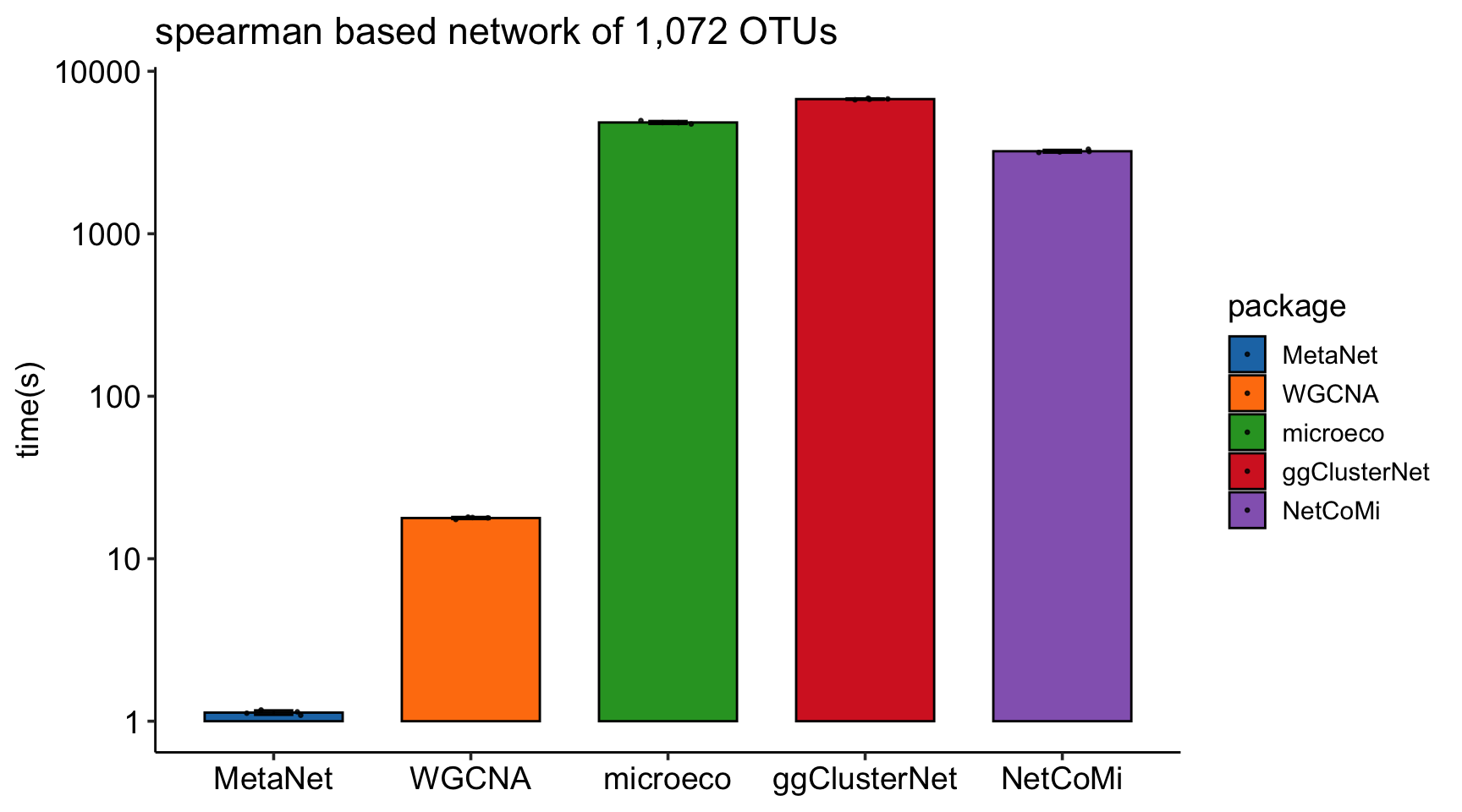
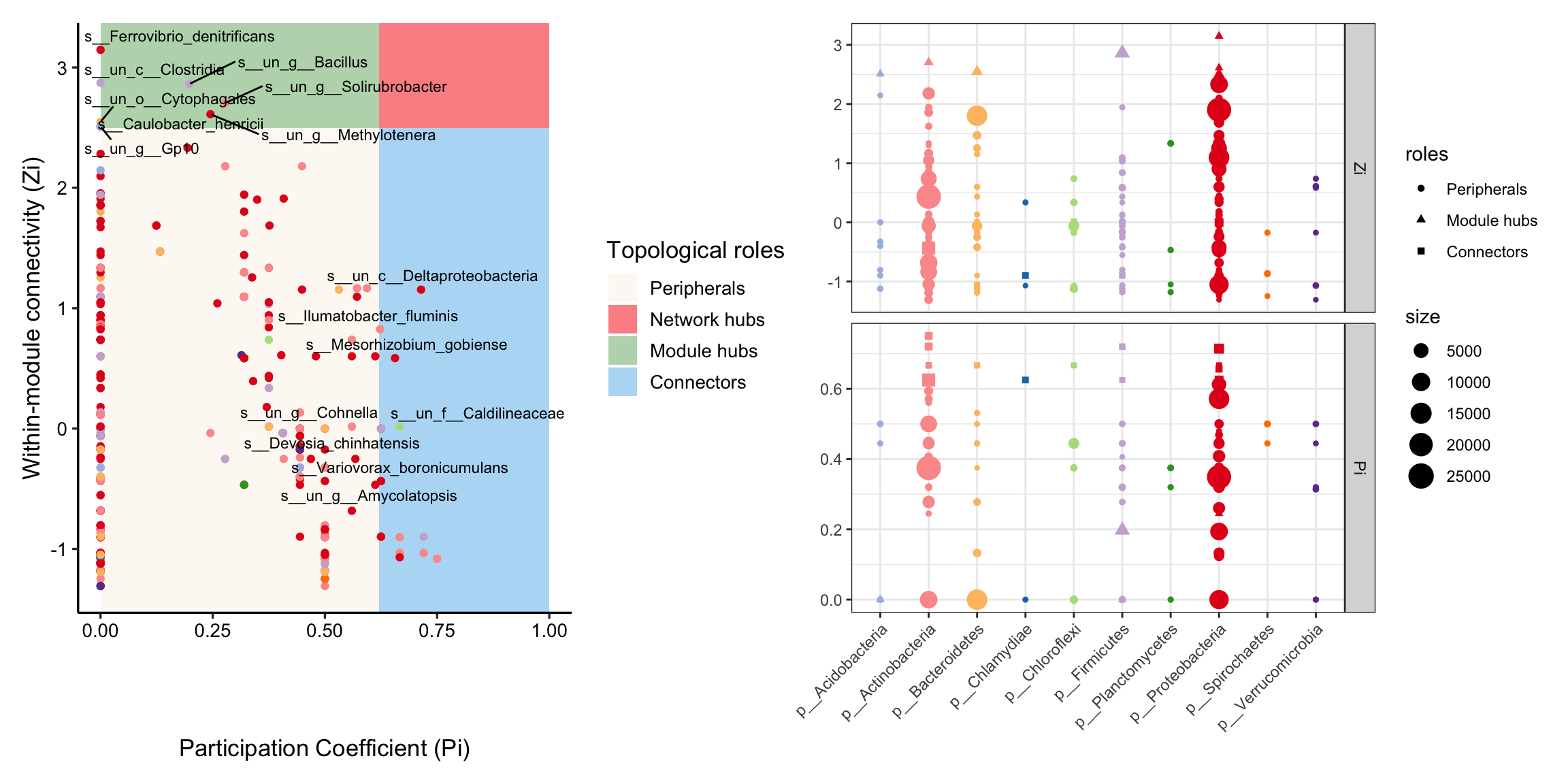
MetaNet features
library(MetaNet)
multi_net_build(list(
Microbiome = micro,
Metabolome = metab,
Transcriptome = transc
)) -> multi1
# set vertex_class
multi1_with_anno <- c_net_set(multi1, micro_g, metab_g, transc_g,
vertex_class = c("Phylum", "kingdom", "type"))
# set vertex_size
multi1_with_anno <- c_net_set(multi1_with_anno,
data.frame("Abundance1" = colSums(micro)),
data.frame("Abundance2" = colSums(metab)),
data.frame("Abundance3" = colSums(transc)),
vertex_size = paste0("Abundance", 1:3)
)
plot(multi1_with_anno)
MetaNet features
MetaNet features

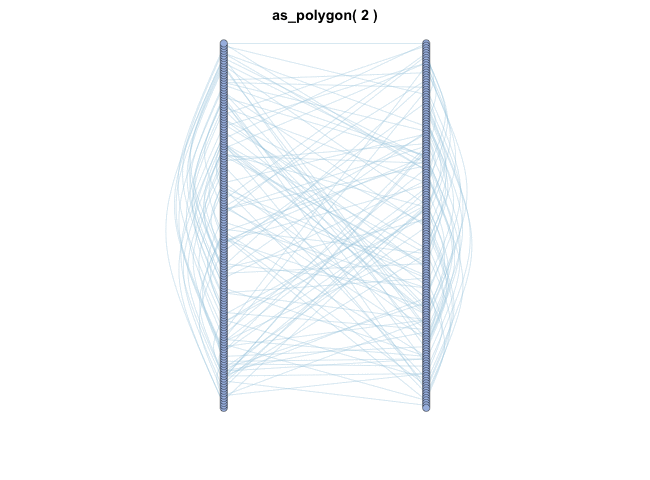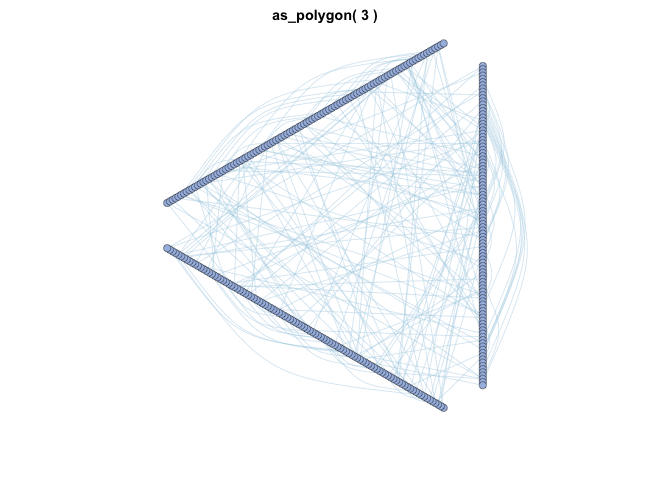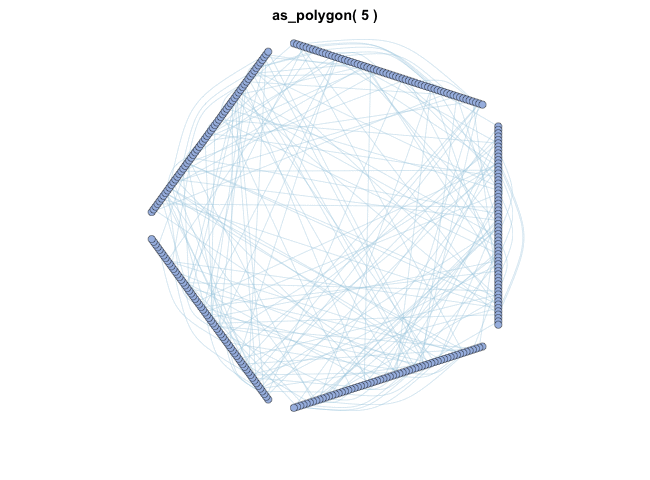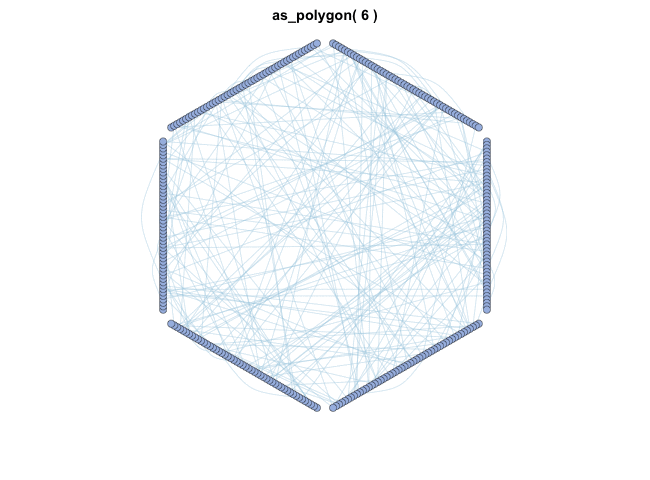

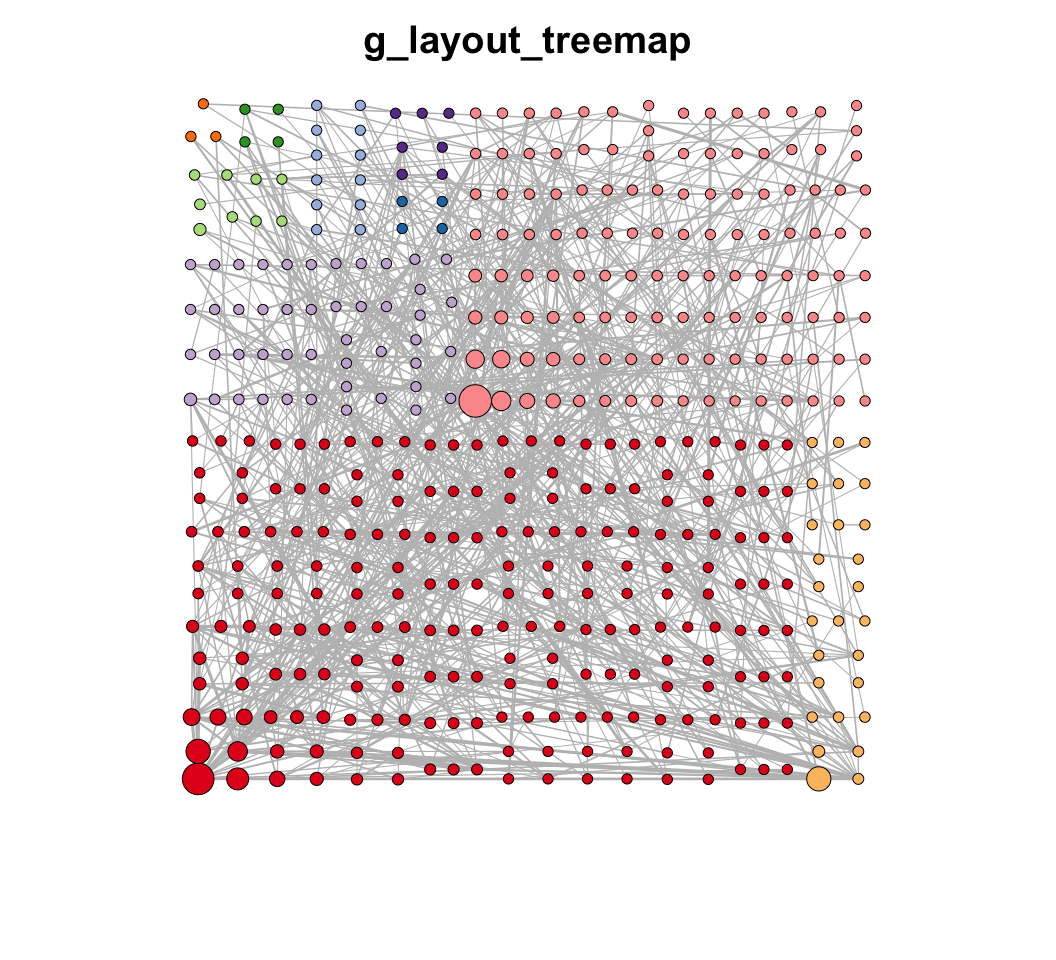
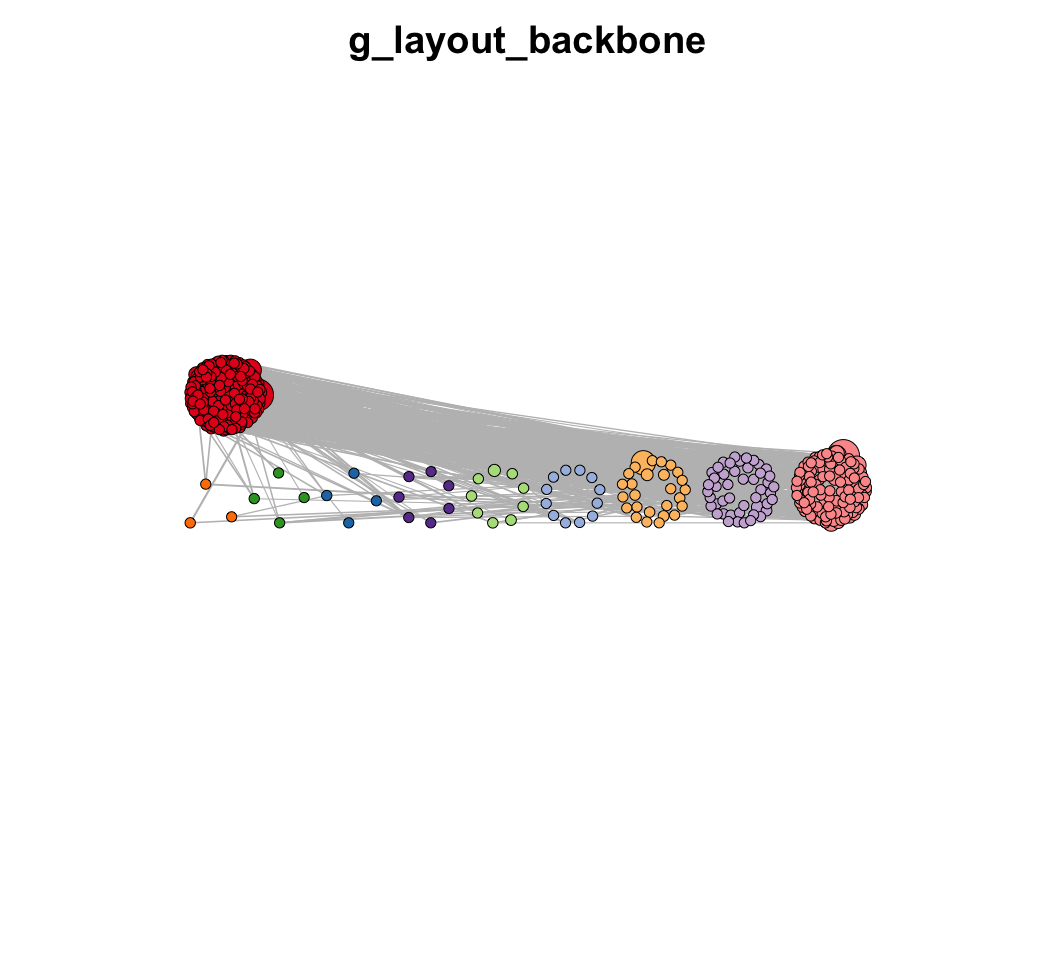

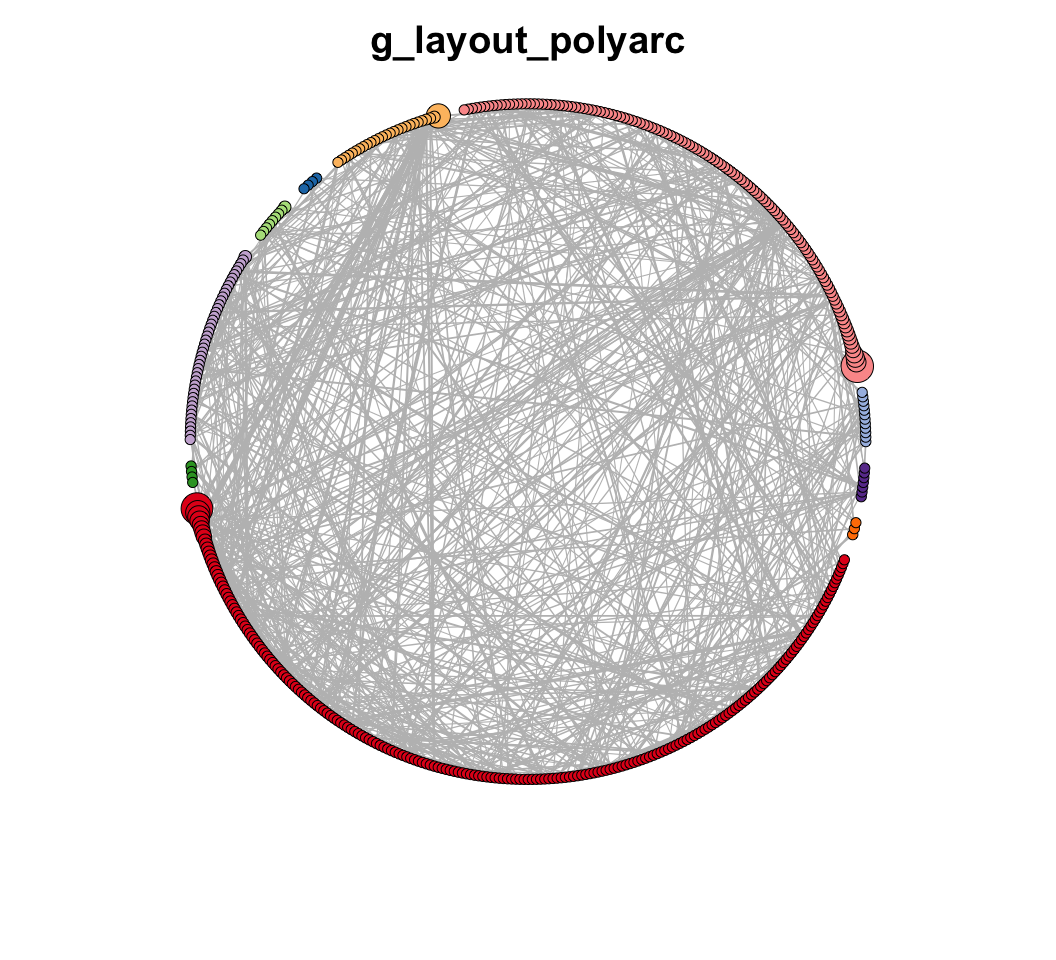
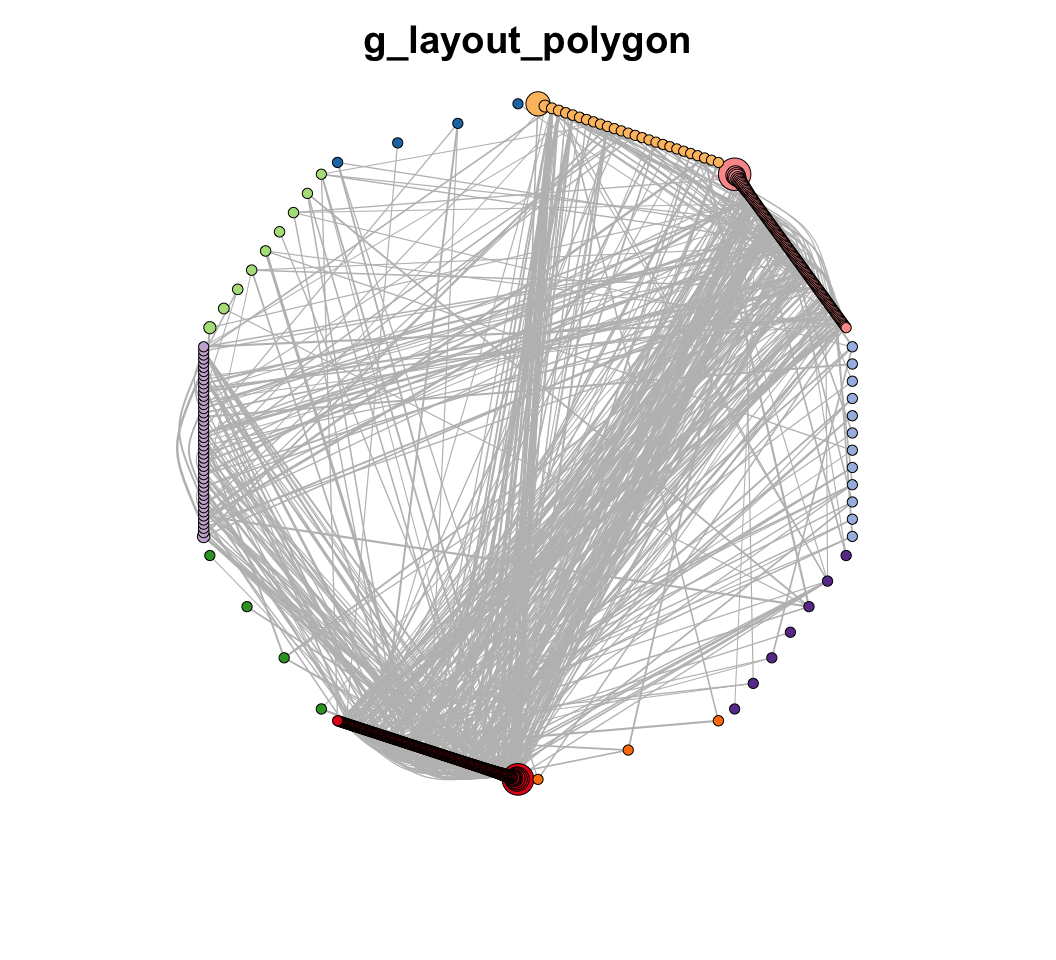
MetaNet features
ReporterScore 
iCRISPR 
CRISPR/Cas9 system analysis in genomics and metagenomics.

plot4fun 
It provides some interesting functions for plotting.
More Resources
Thanks for your attention! 🙏

Asa’s R packages 📦 for bioinformatics and data analysis Chen Peng ZJU LSI Jianglab 2024-04-19