pctax
pctax provides a comprehensive suite of tools for analyzing microbiome data.
Install
|
|
Usage
It includes functionalities for α-diversity analysis, β-diversity analysis, differential analysis, community assembly, visualization of phylogenetic tree and functional enrichment analysis…
Look at the test data:
|
|
α-diversity analysis
Calculate a_diversity of otutab then link to experiment group or environment variable.
|
|
β-diversity analysis
There are a range of dimensionality reduction methods available for analysis, including Constrained and non-Constrained.
Like PCA, PCoA, NMDS, RDA, CCA… For example:
PCA:
|
|
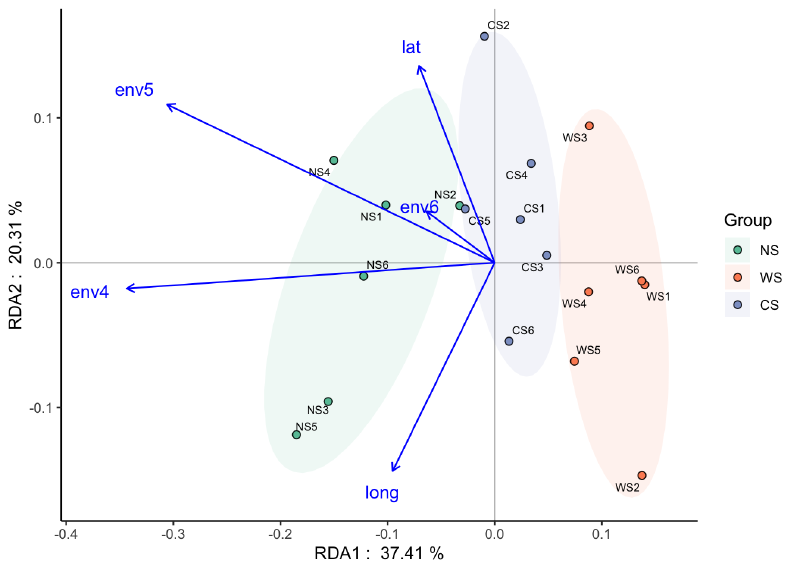
RDA:
|
|
Differential analysis
There are also lots of statistic methods for differential analysis: ALDEX, ANCOM2, randomForest, t.test, wilcox.test… or deseq2, limma…(Commonly used in transcriptome)
|
|
Community assembly
Community assembly in microbiome refers to the processes that shape the composition, diversity, and structure of microbial communities in a particular environment or host. Microbiome consist of diverse microbial populations that interact with each other and their surroundings, and understanding how these communities assemble is crucial for comprehending their ecological dynamics and functional implications.
|
|
Phylogenetic tree
|
|
Cite
Please cite:
Chen P (2023). pctax: Professional Comprehensive Microbiome Data Analysis. R package, https://github.com/Asa12138/pctax.