上一篇推文介绍了MetaNet进行网络可视化以及各种布局方法,本文将介绍一些扩展绘图方法以及兼容工具。
可以从 CRAN 安装稳定版:install.packages("MetaNet")
依赖包 pcutils和igraph(需提前安装),推荐配合 dplyr 进行数据操作。
1
2
3
4
5
6
library ( MetaNet )
library ( igraph )
# ========data manipulation
library ( dplyr )
library ( pcutils )
Copy 兼容工具
ggplot2风格
如果你更熟悉ggplot2,可以使用as.ggig()函数将基础R绘图转换为ggplot2风格:
1
2
as.ggig ( multi1_with_anno ) -> ggig
class ( ggig )
Copy 这样你就可以使用labs()、theme()、ggsave()和cowplot::plot_grid()等函数来制作更好的图形。
Gephi集成
处理大型数据集时,我推荐使用Gephi进行布局,运行比较快且美观。MetaNet提供了通过graphml格式文件与Gephi的接口:
1
c_net_save ( co_net , filename = "~/Desktop/test" , format = "graphml" )
Copy 将test.graphml导入Gephi并进行布局,从Gephi导出graphml文件:test2.graphml,然后在MetaNet中重新绘制:
1
2
3
4
input_gephi ( "~/Desktop/test2.graphml" ) -> gephi
c_net_plot ( gephi $ go , coors = gephi $ coors ,
legend_number = T ,
group_legend_title = "Phylum" )
Copy Cytoscape集成
Cytoscape是另一个优秀的网络可视化软件,包含许多插件。将test.graphml导入Cytoscape进行布局,然后导出为cyjs文件,MetaNet可以读取:
1
2
3
4
input_cytoscape ( "~/Desktop/test2.cyjs" ) -> cyto
c_net_plot ( co_net , coors = cyto $ coors ,
legend_number = T ,
group_legend_title = "Phylum" )
Copy NetworkD3交互式可视化
NetworkD3可以生成基于JavaScript的交互式网络图,输出对象是适合网页的htmlwidgets:
1
netD3plot ( multi1_with_anno )
Copy 扩展绘图
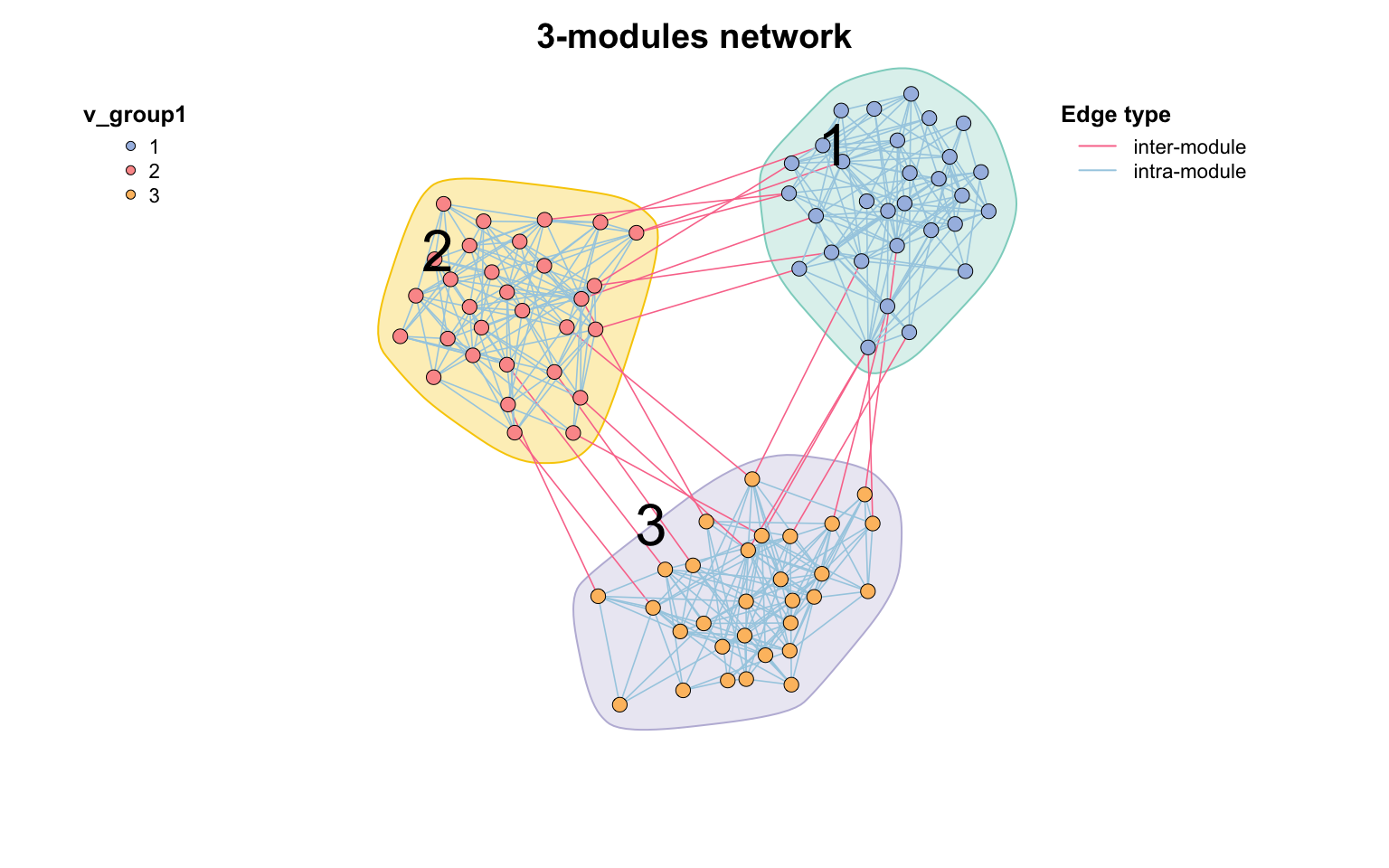
模块图
使用mark_module=T在网络中标记你的模块,下次会具体介绍网络模块分析:
1
2
test_module_net <- module_net ( module_number = 3 , n_node_in_module = 30 )
c_net_plot ( test_module_net , mark_module = T , module_label = T , module_label_just = c ( 0.2 , 0.8 ))
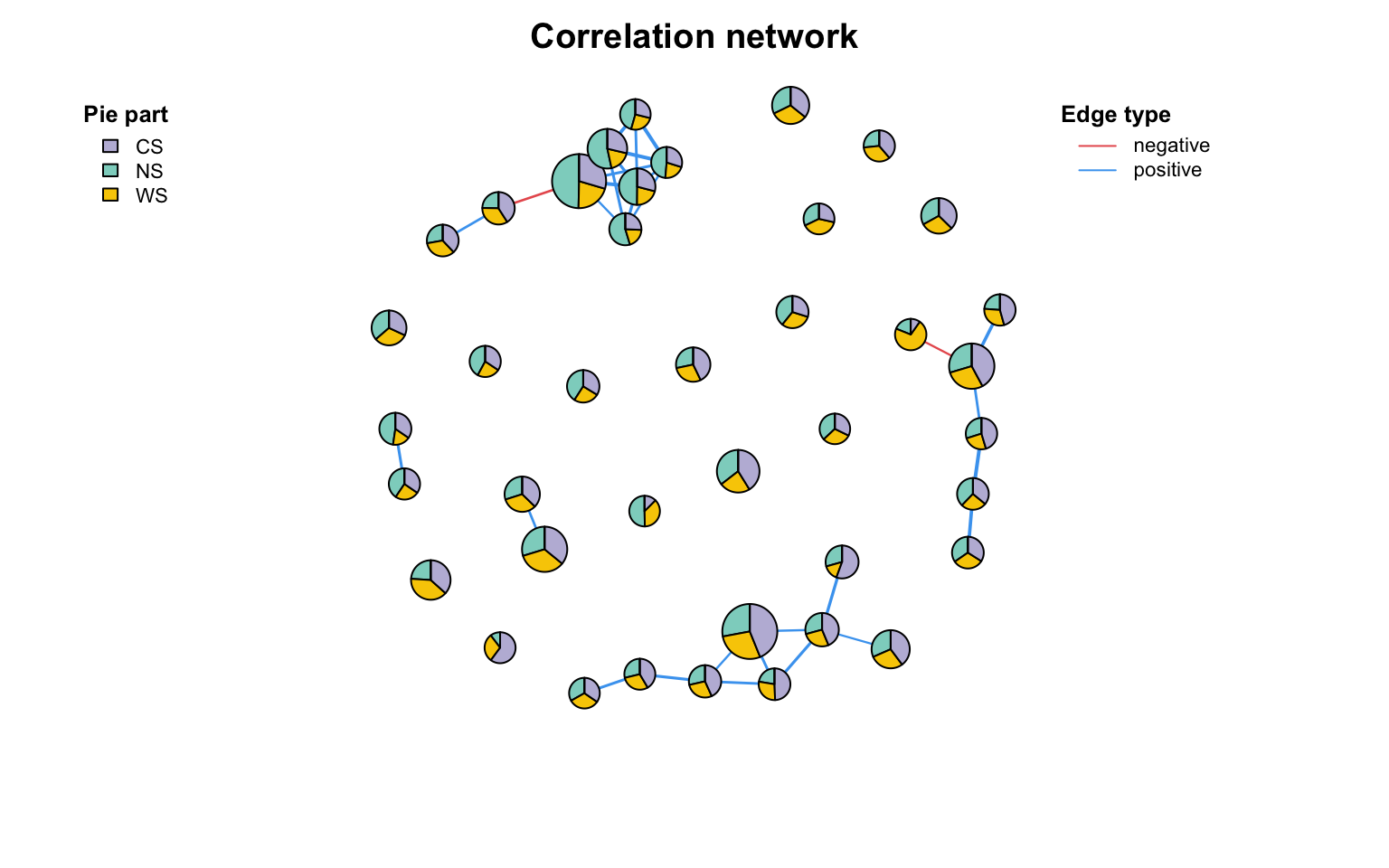
Copy pie节点
把网络里的节点直接画成pie图,不用在AI里面一个个拖过去了:
1
2
3
4
data ( "otutab" )
data ( "c_net" )
hebing ( otutab , metadata $ Group ) -> otutab_G
head ( otutab_G )
Copy
1
2
3
4
5
6
7
## NS WS CS
## s__un_f__Thermomonosporaceae 1218.3333 1227.3333 1912.1667
## s__Pelomonas_puraquae 2087.6667 873.5000 1241.6667
## s__Rhizobacter_bergeniae 819.1667 781.3333 1164.8333
## s__Flavobacterium_terrae 816.1667 944.6667 986.5000
## s__un_g__Rhizobacter 821.8333 540.6667 953.3333
## s__un_o__Burkholderiales 972.6667 327.5000 522.6667
Copy
1
2
3
4
co_net_f = c_net_filter ( co_net , name %in% head ( rownames ( otutab_G ), 40 ))
c_net_plot ( co_net_f , pie_value = otutab_G ,
vertex.shape = c ( "pie" ), #把你需要展示pie的节点的shape设置为pie
pie_legend = T , color_legend = F , vertex_size_range = c ( 10 , 18 ))
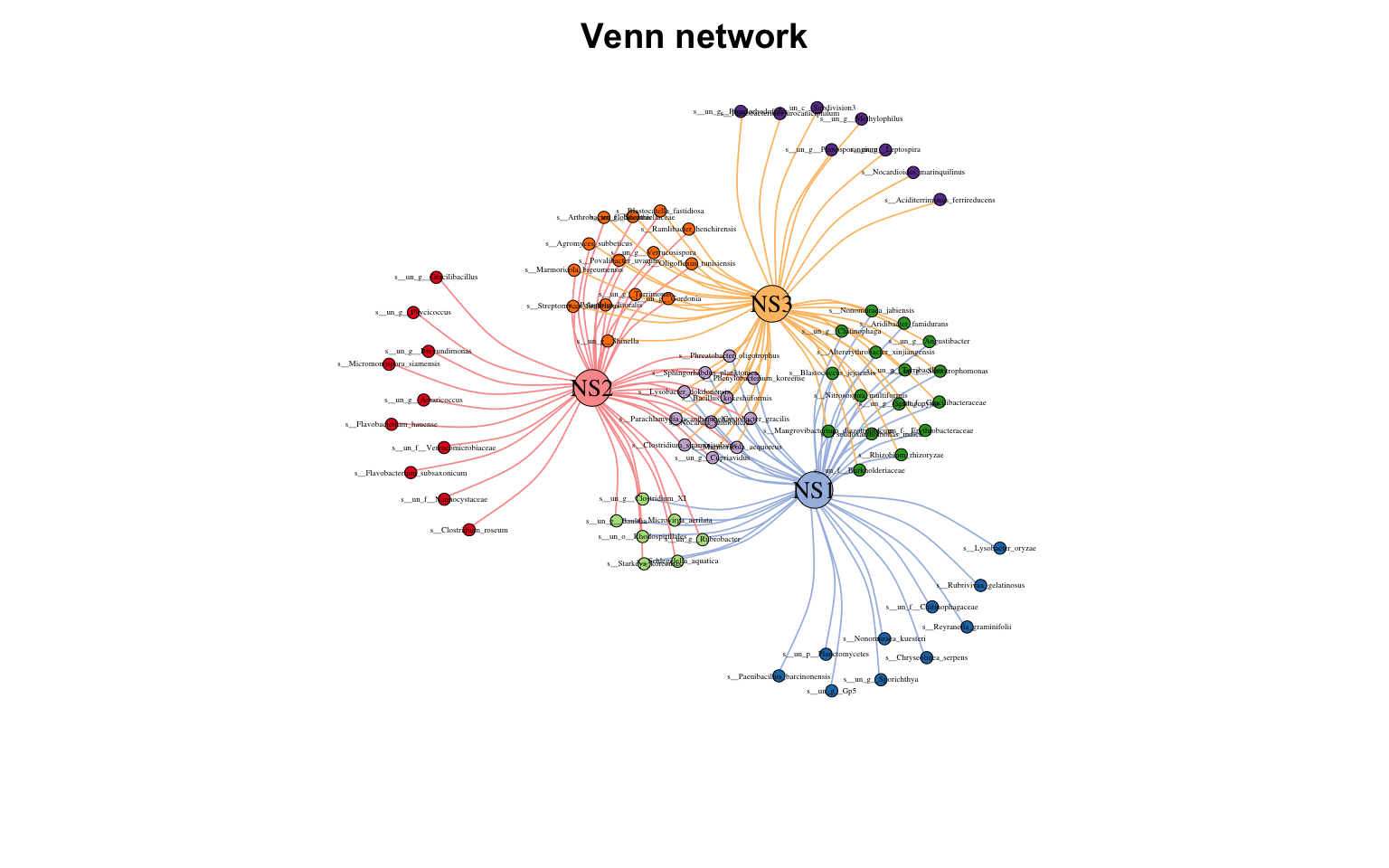
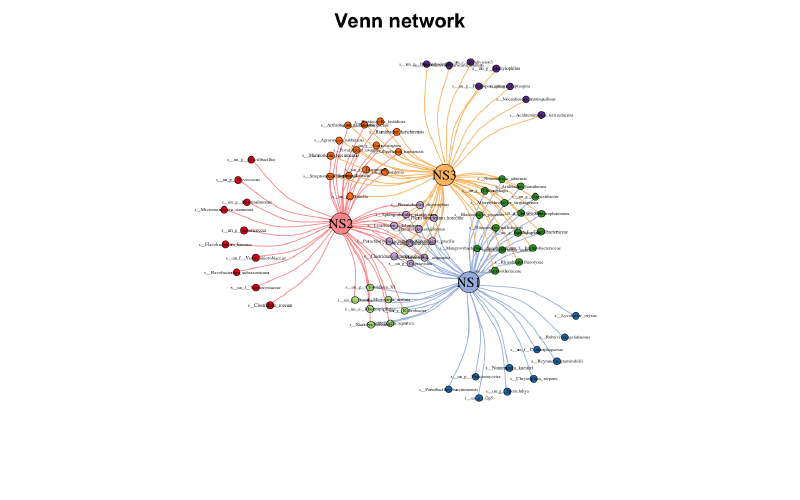
Copy 韦恩图网络
当集合里的点数量合适时,韦恩图网络算是个不错的展示方法
1
2
3
4
data ( otutab , package = "pcutils" )
tab <- otutab[400 : 485 , 1 : 3 ]
head ( tab ) #0代表不存在
Copy
1
2
3
4
5
6
7
## NS1 NS2 NS3
## s__un_g__Brevundimonas 0 4 0
## s__un_g__Shinella 0 10 12
## s__Ramlibacter_henchirensis 0 9 6
## s__Cystobacter_gracilis 3 4 17
## s__un_g__Gordonia 0 6 16
## s__un_f__Nannocystaceae 0 6 0
Copy
1
2
venn_net ( tab ) -> v_net
plot ( v_net )
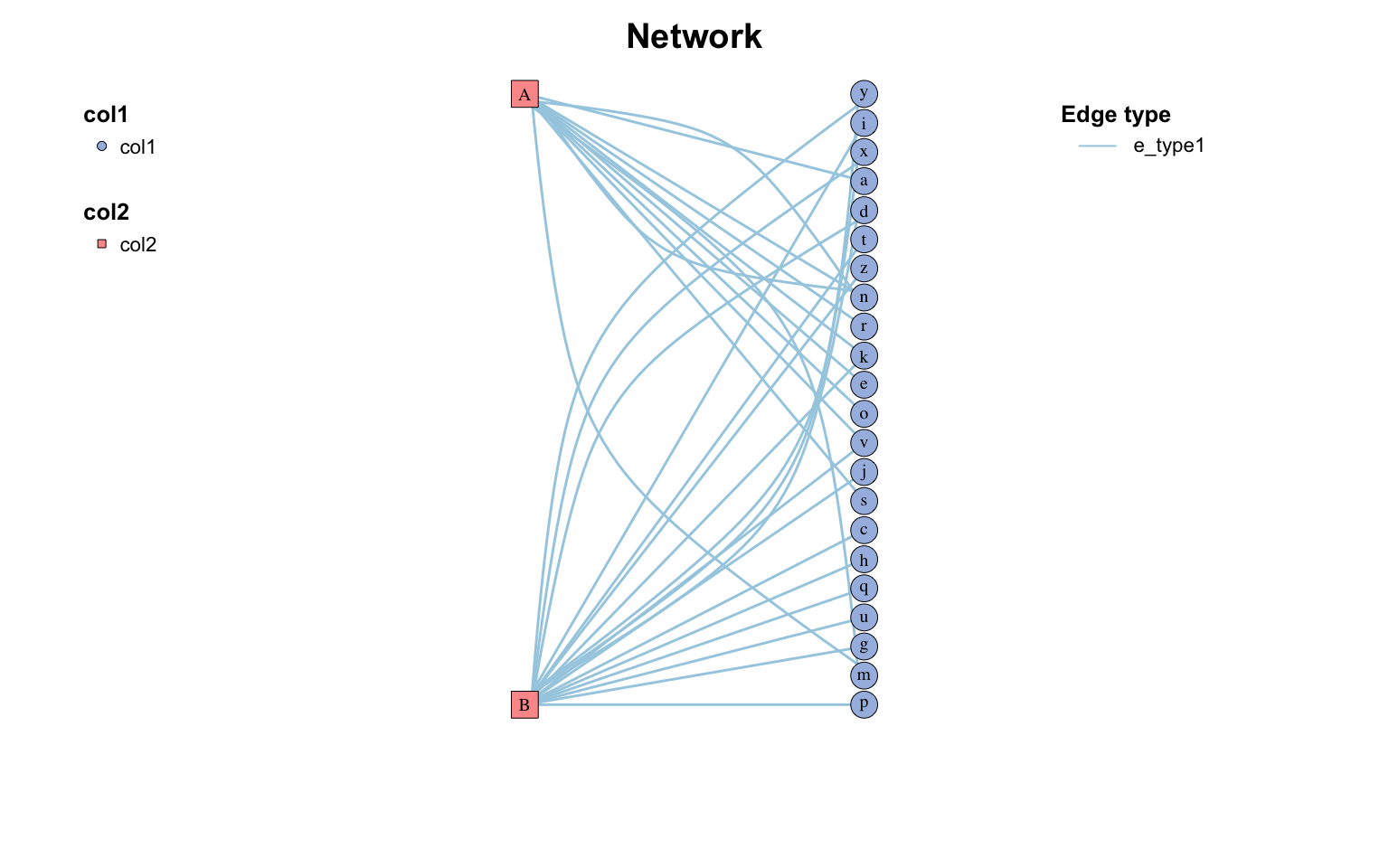
Copy 两列边列表
1
2
3
4
5
6
twocol <- data.frame (
"col1" = sample ( letters , 30 , replace = TRUE ),
"col2" = sample ( c ( "A" , "B" ), 30 , replace = TRUE )
)
twocol_net <- twocol_edgelist ( twocol )
c_net_plot ( twocol_net , g_layout_polygon ( twocol_net ), labels_num = "all" )
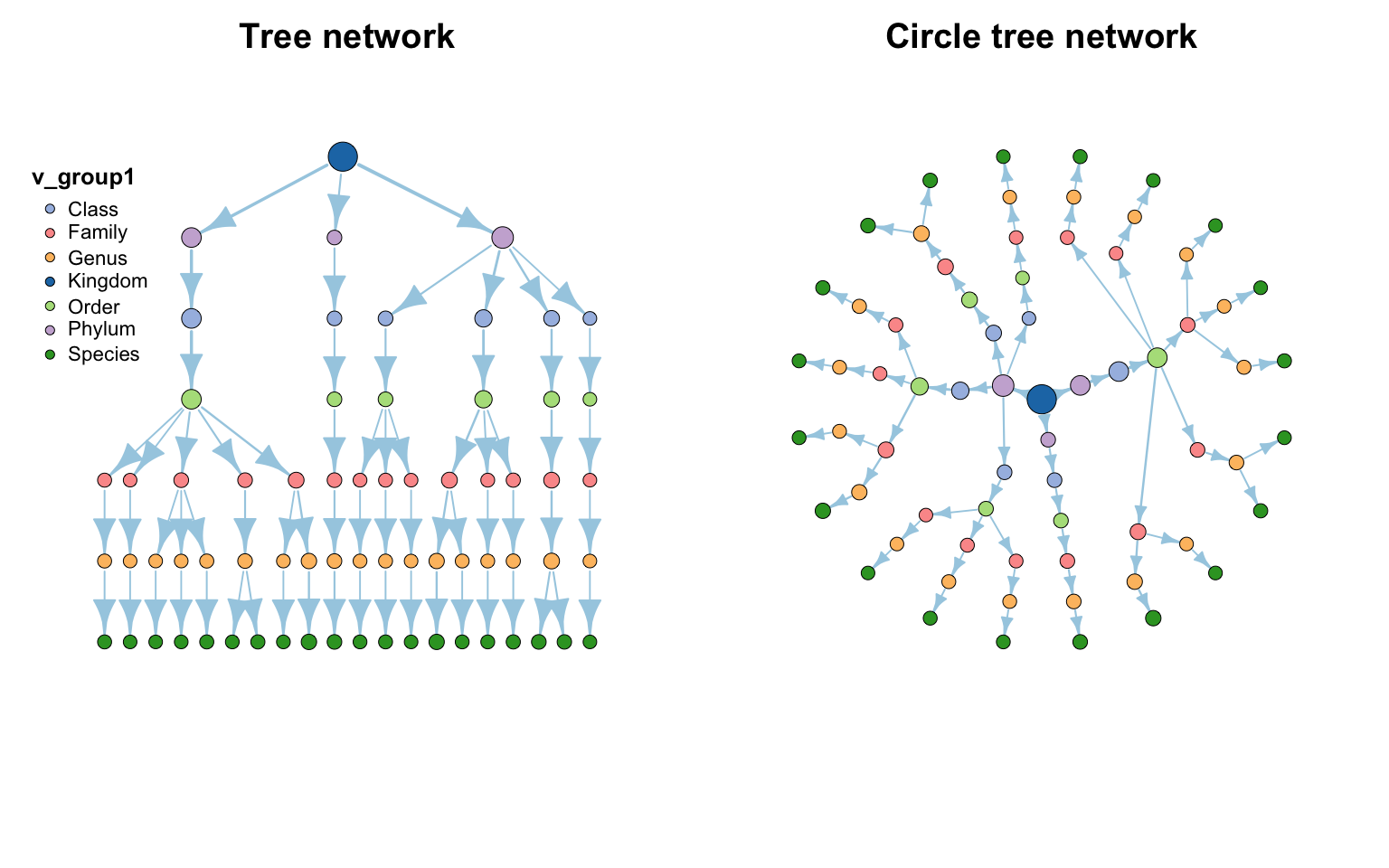
Copy 网络树
1
2
3
4
5
6
7
8
9
data ( "otutab" , package = "pcutils" )
cbind ( taxonomy , num = rowSums ( otutab )) [1 : 20 , ] -> test
df2net_tree ( test ) -> ttt
par ( mfrow = c ( 1 , 2 ))
plot ( ttt , edge_legend = F , main = "Tree network" , legend_position = c ( left_leg_x = -1.3 ),
edge.arrow.size = 1 , edge.arrow.width = 1 , rescale = T )
plot ( ttt , coors = as_circle_tree (), legend = F , main = "Circle tree network" ,
edge.arrow.size = 0.5 , edge.arrow.width = 1 , rescale = T )
Copy 奥运五环图
这个也是上次讲的分组布局的应用了,g_layout函数布局,用下面的代码就可以绘制:
1
2
3
4
5
6
7
8
9
10
11
12
13
r <- 1
pensize <- r / 6
rings_data <- data.frame (
x = c ( -2 * ( r + pensize ), - ( r + pensize ), 0 , ( r + pensize ), 2 * ( r + pensize )),
y = c ( r , 0 , r , 0 , r ),
color = c ( "#0081C8" , "#FCB131" , "#000000" , "#00A651" , "#EE334E" )
)
g1 <- module_net ( module_number = 5 , n_node_in_module = 30 )
plot ( g1 ,
coors = g_layout ( g1 , layout1 = rings_data[ , 1 : 2 ] , zoom1 = 1.2 , zoom2 = 0.5 ),
rescale = FALSE , legend = FALSE , main = "Olympic Rings" , vertex.frame.color = NA ,
edge.width = 0 , vertex.color = setNames ( rings_data $ color , 1 : 5 ), vertex.size = 7
)
Copy References
Koutrouli M, Karatzas E, Paez-Espino D and Pavlopoulos GA (2020) A Guide to Conquer the Biological Network Era Using Graph Theory. Front. Bioeng. Biotechnol. 8:34. doi: 10.3389/fbioe.2020.00034
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. https://doi.org/10.1038/nrmicro2832 .
Y. Deng, Y. Jiang, Y. Yang, Z. He, et al., Molecular ecological network analyses. BMC bioinformatics (2012), doi:10.1186/1471-2105-13-113.